
How to Perform ANOVA with Python
Important: This blog post shows you how to perform ANOVA and check its assumptions using Python. If you’re looking for a general introduction, check out the related blog posts linked below.
In earlier posts, we explored the theoretical foundations of ANOVA and model building in DOE. We covered how to build mathematical models, systematic model building with ANOVA, and how to check ANOVA assumptions. Now, let’s dive into the practical side and learn how to perform these analyses using Python.
Python is a favorite for data analysis, and it’s easy to see why. It’s free, powerful, and packed with excellent statistical libraries. Even if you’re not a programming expert, tools like ChatGPT and modern AI assistants make it easier than ever to get started.
Let’s walk through the process step by step, using our familiar filtration rate example. I’ll show you how to build the model in Python and validate it by checking ANOVA assumptions.
Setting Up Your Python Environment
First, install the required packages. If you’re using Anaconda (recommended for beginners), most of these come pre-installed:
# Required imports
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm
from statsmodels.formula.api import ols
from scipy.stats import shapiroIf you need to install any packages, use:
pip install package_nameYour First ANOVA with Python
Main Effects
Now let’s perform a basic ANOVA using the forward selection approach we learned about. We’ll start with the main effects of our experiment:
# Read data
df = pd.read_excel("filtration_rate_example.xlsx")
# Define the formula for the model
formula = 'Filtration_rate ~ T + P + CoF + RPM'
# Fit the model
model = ols(formula, data=df).fit()
# Perform ANOVA
anova_results = sm.stats.anova_lm(model)
# Print the ANOVA results
print(anova_results)This produces an ANOVA table similar to what we discussed in theory:
Output:
df sum_sq mean_sq F PR(>F)
T 1.0 1870.5625 1870.562500 7.988620 0.016478
P 1.0 39.0625 39.062500 0.166824 0.690788
CoF 1.0 390.0625 390.062500 1.665842 0.223284
RPM 1.0 855.5625 855.562500 3.653855 0.082330
Residual 11.0 2575.6875 234.153409 NaN NaNUnderstanding the ANOVA table columns:
- df: Degrees of freedom - the number of independent pieces of information for each source
- sum_sq: Sum of squares - the total variation attributed to each factor
- mean_sq: Mean square - sum of squares divided by degrees of freedom (variance estimate)
- F: F-statistic - the ratio of factor variance to residual variance
- PR(>F): P-value - probability of seeing this F-statistic by chance alone
Adding Interactions
But we’re still missing the two important interactions that we identified in our previous analysis. Let’s add the T×CoF and T×RPM interactions to our model:
# Updated formula with interactions
# The ':' symbol creates interaction terms
formula_with_interactions = 'Filtration_rate ~ T + P + CoF + RPM + T:CoF + T:RPM'
# Fit the new model
model_interactions = ols(formula_with_interactions, data=df).fit()
# Perform ANOVA
anova_interactions = sm.stats.anova_lm(model_interactions)
print(anova_interactions)Understanding the interaction syntax:
T:CoFcreates the Temperature × Concentration Factor interactionT:RPMcreates the Temperature × RPM interaction- You can also use
*as shorthand:T*CoFexpands toT + CoF + T:CoF
The updated ANOVA table now includes our interactions:
Output:
df sum_sq mean_sq F PR(>F)
T 1.0 1870.5625 1870.562500 107.873849 0.000003
P 1.0 39.0625 39.062500 2.252703 0.167622
CoF 1.0 390.0625 390.062500 22.494594 0.001055
RPM 1.0 855.5625 855.562500 49.339608 0.000062
T:RPM 1.0 1105.5625 1105.562500 63.756908 0.000022
T:CoF 1.0 1314.0625 1314.062500 75.780937 0.000011
Residual 9.0 156.0625 17.340278 NaN NaNFor the final model, we also remove the main effect of pressure since it’s insignificant.
# Updated formula with interactions
# The ':' symbol creates interaction terms
formula_with_interactions = 'Filtration_rate ~ T + CoF + RPM + T:CoF + T:RPM'
# Fit the new model
model_interactions = ols(formula_with_interactions, data=df).fit()
# Perform ANOVA
anova_interactions = sm.stats.anova_lm(model_interactions)
print(anova_interactions)Output:
df sum_sq mean_sq F PR(>F)
T 1.0 1870.5625 1870.5625 95.864830 0.000002
CoF 1.0 390.0625 390.0625 19.990391 0.001195
RPM 1.0 855.5625 855.5625 43.846893 0.000059
T:RPM 1.0 1105.5625 1105.5625 56.659193 0.000020
T:CoF 1.0 1314.0625 1314.0625 67.344651 0.000009
Residual 10.0 195.1250 19.5125 NaN NaNAssumption Checking with Python
Now that we have performed the ANOVA, we need to check the three ANOVA assumptions:
- Normality
- Independence
- Equal variance
Extracting Residuals
All three assumption tests require analyzing the residuals, so let’s extract them along with the fitted values.
# Extract residuals and fitted values from our final model
residuals = model_interactions.resid
fitted = model_interactions.fittedvaluesUnderstanding the code syntax:
model_interactions.residextracts the residuals (the difference between observed values and model predictions)model_interactions.fittedvaluesextracts the fitted values (the values predicted by the model for each experimental run)
These two arrays contain the core information we need to validate our ANOVA assumptions.
residuals:
0 -1.250
1 1.625
2 1.750
3 -4.375
4 -6.250
5 -1.125
6 5.750
7 3.875
8 -1.250
9 -0.625
10 0.750
11 3.375
12 2.750
13 -6.375
14 -2.250
15 3.625
dtype: float64fitted values:
0 46.250
1 69.375
2 46.250
3 69.375
4 74.250
5 61.125
6 74.250
7 61.125
8 44.250
9 100.625
10 44.250
11 100.625
12 72.250
13 92.375
14 72.250
15 92.375
dtype: float64Testing Normality
To test whether the residuals follow a normal distribution, we’ll use a Q-Q plot. This is important because ANOVA’s statistical tests (like p-values) assume normality. If the residuals aren’t normal, the p-values might be unreliable.
# Q-Q plot of residuals
sm.qqplot(residuals, line="s")
plt.title("Q-Q Plot of ANOVA Residuals")
plt.show()Code explanation:
sm.qqplot()creates a quantile-quantile plot comparing residuals to a theoretical normal distributionline="s"adds a reference line to help visualize normality- If the points fall roughly along the line, the normality assumption is satisfied
The output will be the following figure:
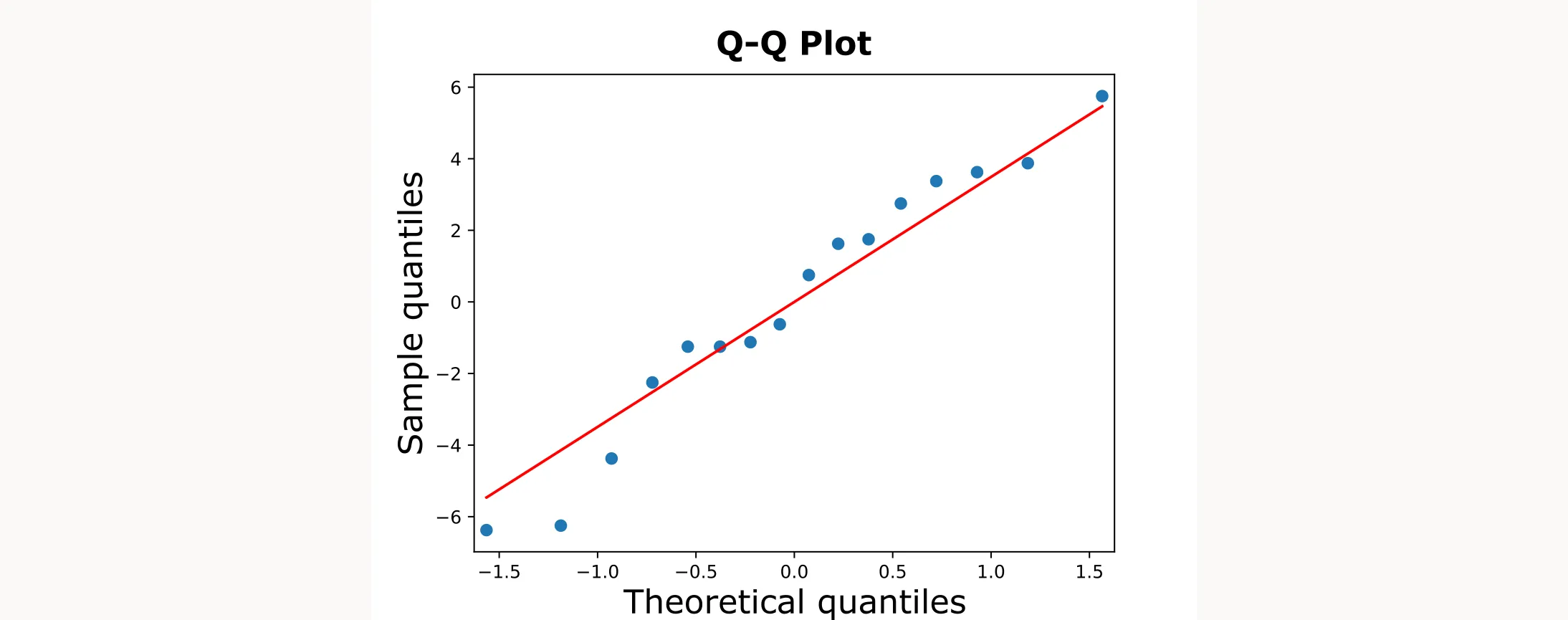
Testing Independence
Next, we’ll test for independence by plotting residuals against the run order.
# Residuals vs run/order (look for patterns/trends)
plt.scatter(df.index, residuals)
plt.xlabel('Run Order')
plt.ylabel('Residuals')
plt.title('Residuals vs Run Order')
plt.axhline(0, color='red', linestyle='--')
plt.savefig("Residuals_vs_RunOrder.svg")
plt.show()Code explanation:
plt.plot(df["order"], residuals, marker="o", linestyle="-")creates a line plot of residuals against run orderplt.axhline(0, color='red', linestyle='--')adds a red, horizontal dashed line at zero as a reference- We’re looking for random scatter around zero. Any trends or patterns suggest the independence assumption is violated
This is how the output figure will look like:
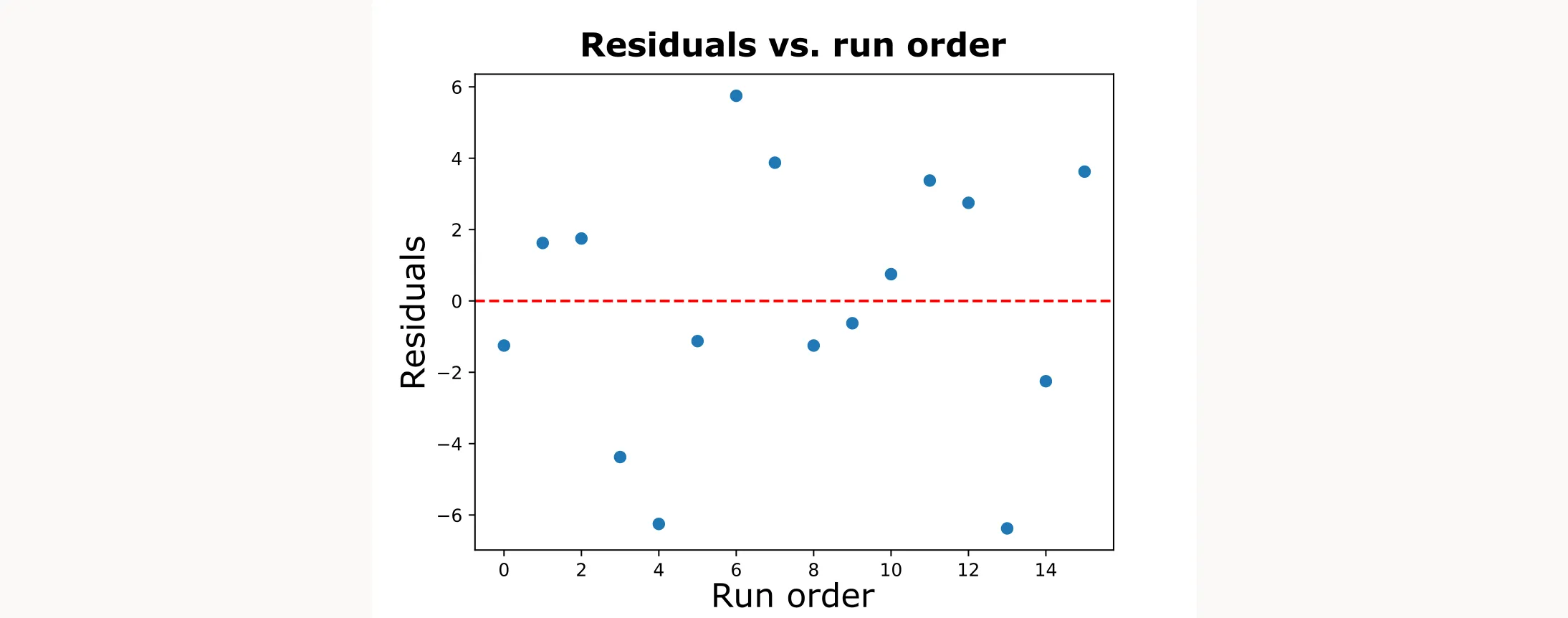
Testing Equal Variance
# Residuals vs fitted values (look for “funnel” shapes)
plt.scatter(fitted, residuals)
plt.axhline(0, color='red', linestyle="--")
plt.title("Residuals vs Fitted")
plt.xlabel("Fitted values")
plt.ylabel("Residuals")
plt.savefig("Residuals_vs_Fitted.svg")
plt.show()Code explanation:
plt.scatter(fitted, residuals)creates a scatter plot of residuals against fitted valuesplt.axhline(0, color='red', linestyle="--")adds a red, horizontal dashed line at zero as a reference- We’re looking for a constant spread of residuals around zero. A “funnel” shape (narrow at one end and wide at the other) indicates a violation of the equal variance assumption
This is how it looks like:
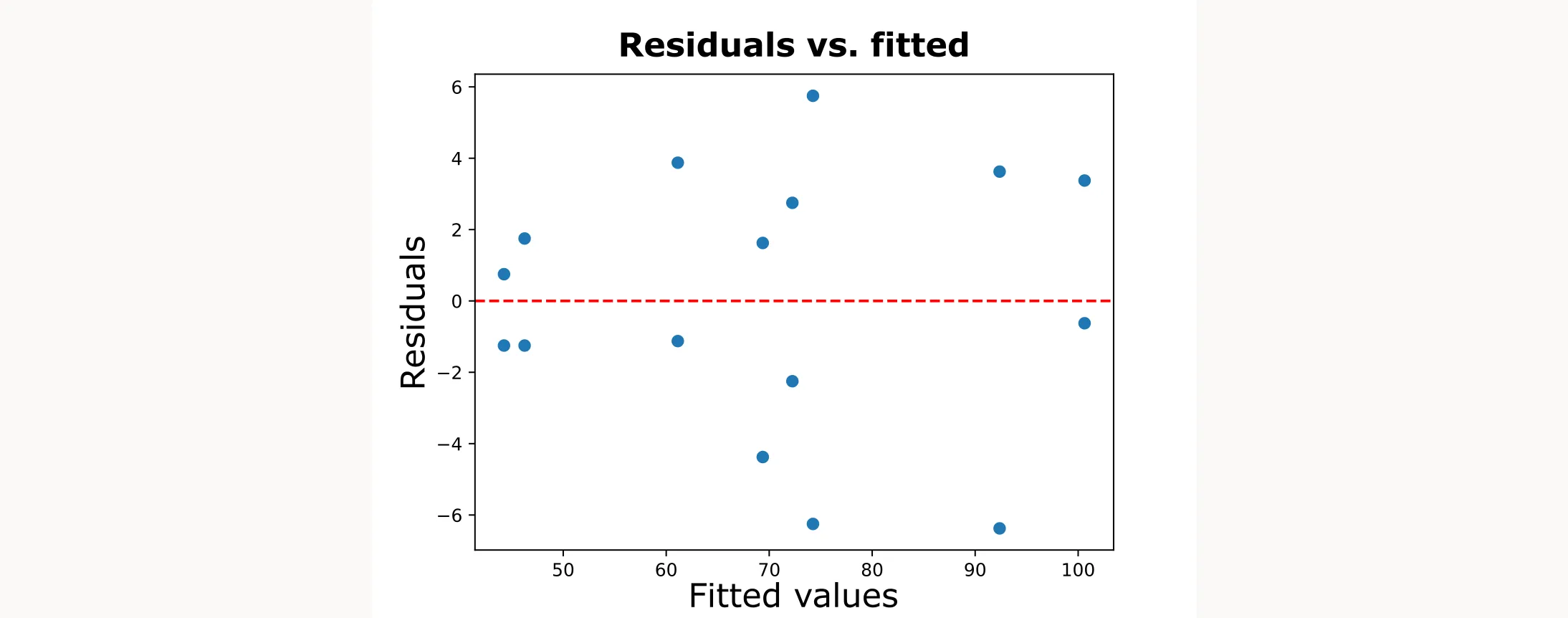
Adding Quadratic Terms for Advanced Designs
If you’re working with a central composite design (CCD) or other response surface methodology, you can add quadratic terms to capture non-linear relationships. The anlysis as well as the testing of ANOVA assumptions will be the same:
# Adding quadratic terms
# Note: I(T**2) syntax tells Python to treat T**2 as a mathematical operation
formula_quad = 'Filtration_rate ~ T + CoF + RPM + T:CoF + T:RPM + I(T**2) + I(CoF**2) + I(RPM**2)'
model_quad = ols(formula_quad, data=df).fit()
anova_results_quad = sm.stats.anova_lm(model_quad, typ=2) # Type II ANOVA
print(anova_results_quad)Important note: You can only meaningfully add quadratic terms if your experimental design includes center points or multiple levels for each factor. A simple two-level factorial design doesn’t provide enough information to estimate quadratic effects reliably.
The I() function: This tells the formula parser to treat the expression inside as a mathematical operation rather than a formula operator. Without it, T**2 might be misinterpreted.